Overview
- GST fusion protein with the sequence ELANRAEVPLSWSVS SKLNQHAELETEEEEKNPEELTERNG, corresponding to residues 374-414 of mouse Kir3.2 (Accession P48542). Intracellular, C-terminal part.

- Mouse brain membrane proteins (1:174) (Jelacic, T.M. et al. (2000) J. Biol. Chem. 275, 36211.).
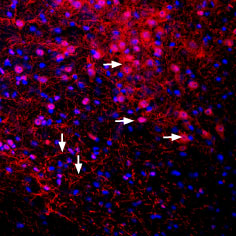 Expression of Kir3.2 in mouse brainImmunohistochemical staining of perfusion-fixed frozen mouse substantia nigra pars compacta sections using Anti-GIRK2 (Kir3.2) Antibody (#APC-006), (1:400). GIRK2 staining (red) appears in cells and processes along the pars compacta of the mouse substantia nigra (horizontal arrows) and in the pars reticulata (vertical arrows). Cell nuclei are stained with DAPI (blue).
Expression of Kir3.2 in mouse brainImmunohistochemical staining of perfusion-fixed frozen mouse substantia nigra pars compacta sections using Anti-GIRK2 (Kir3.2) Antibody (#APC-006), (1:400). GIRK2 staining (red) appears in cells and processes along the pars compacta of the mouse substantia nigra (horizontal arrows) and in the pars reticulata (vertical arrows). Cell nuclei are stained with DAPI (blue).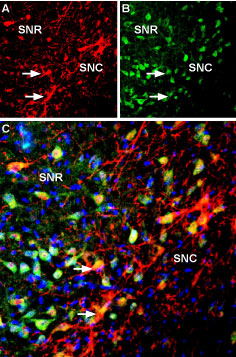 Multiplex staining of Kir3.2 and KCNK2 in rat substantia nigraImmunohistochemical staining of immersion-fixed, free floating rat brain frozen sections using rabbit Anti-GIRK2 (Kir3.2) Antibody (#APC-006), (1:400) and Guinea pig Anti-KCNK2 (TREK-1) Antibody (#APC-047-GP), (1:120). A. Kir3.2 staining (red) appears in cells of the substantia nigra pars compacta (SNC, arrows). B. KCNK2 (green) appears in both compacta (SNC) and reticulata (SNR) portions of the substantia nigra. C. Merge of the two images reveals co-localization in some cells (arrows), mainly in the SNC region. Cell nuclei are stained with DAPI (blue).
Multiplex staining of Kir3.2 and KCNK2 in rat substantia nigraImmunohistochemical staining of immersion-fixed, free floating rat brain frozen sections using rabbit Anti-GIRK2 (Kir3.2) Antibody (#APC-006), (1:400) and Guinea pig Anti-KCNK2 (TREK-1) Antibody (#APC-047-GP), (1:120). A. Kir3.2 staining (red) appears in cells of the substantia nigra pars compacta (SNC, arrows). B. KCNK2 (green) appears in both compacta (SNC) and reticulata (SNR) portions of the substantia nigra. C. Merge of the two images reveals co-localization in some cells (arrows), mainly in the SNC region. Cell nuclei are stained with DAPI (blue).- Mouse spinal cords (Marker, C.L. et al. (2004) J. Neurosci. 24, 2806.).
- hVM1 (human ventral mesencephalic neural stem cell line 1 (1:400) (Courtois, E.T. et al. (2010) J. Biol. Chem. 285, 9881.).
- Hess, E.J. (1996) Neuron 16, 1073.
- Dascal, N. (1997) Cell Signal. 9, 551.
- Mark, M.D. et al. (2000) Eur. J. Biochem. 267, 5830.
- Kubo, Y. et al. (2005) Pharmacol. Rev. 57, 509.
Kir3.2 (or G-protein regulated inward-rectifier K+ channel, GIRK2) is a member of the family of inward rectifying K+ channels. The family includes 15 members that are structurally and functionally different from the voltage-dependent K+ channels.
The family’s topology consists of two transmembrane domains that flank a single and highly conserved pore region with intracellular N- and C-termini. As is the case for the voltage-dependent K+ channels, the functional unit for the Kir channels is composed of four subunits that can assemble as either homo- or heterotetramers.
Kir channels are characterized by a K+ efflux that is limited by depolarizing membrane potentials thus making them essential for controlling resting membrane potential and K+ homeostasis.
Kir3.2 is a member of the Kir3.x subfamily that includes four members (Kir3.1- Kir3.4). The Kir3 family is characterized by the fact that the channels can be activated by neurotransmitters and other factors acting via the activation of G-protein coupled receptors. Binding of the corresponding ligand to the G-protein receptor induces the dissociation of Gα-GTP from the Gbg dimer. The latter directly binds to Kir3 and activates the channel.2,3
Kir3.2 is mainly expressed in the brain, where it co-assembles with Kir3.1 (GIRK1) or Kir3.3 (GIRK3) and mediates the inhibitory effects of many neurotransmitters including opioid, adrenergic, muscarinic, dopaminergic and GABAergic neurotransmitters.2,3
Point mutations in the mouse Kir3.2 channel cause the weaver (wv) phenotype, a neurological abnormality characterized by the abnormal ‘weaving’ of the mice when they walk, hence the name weaver which is due to a substantial loss of cerebellar granule neurons. These mice also display mild local motor hyperactivity, presumably caused by the degeneration of dopaminergic neurons in the substantia nigra, spontaneous seizures and male sterility.1
A peptide toxin originating from the Apis mellifera bee venom, Tertiapin (#STT-250) was shown to be a potent blocker of Kir3.2 containing channels (7 nM for Kir3.2 alone and 5.4 nM for the Kir3.1/3.2 combination).4
Application key:
Species reactivity key:
Anti-GIRK2 (Kir3.2) Antibody (#APC-006) is a highly specific antibody directed against an epitope of the mouse protein. The antibody can be used in western blot, immunoprecipitation, immunocytochemistry, and immunohistochemistry applications. It has been designed to recognize Kir3.2 from rat, mouse, and human samples.
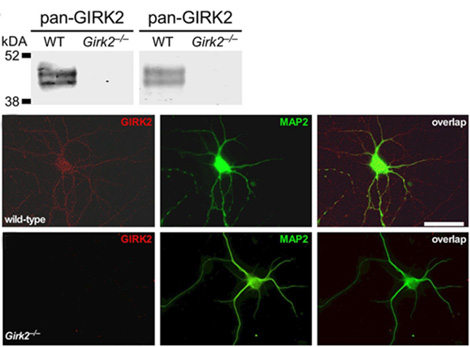
Knockout validation of Anti-GIRK2 (Kir3.2) Antibody in mouse hippocampus.Western blot analysis of mouse hippocampus lysate using Anti-GIRK2 (Kir3.2) Antibody (#APC-006) (upper panels). Note lack of GIRK2 in knockout mice. Immunocytochemical staining of cultured hippocampal mouse neurons with Anti-GIRK2 (Kir3.2) Antibody in wild type cells (upper staining) and Girk2-/- cells (lower staining).Adapted from Marron Fernandez de Velasco, E. et al. (2017) Sci. Rep. 7, 1639. with permission of SPRINGER NATURE.
Applications
Citations
- Immunohistochemical staining of mouse brain sections (1:400). Tested in GIRK2 conditional knockout mice.
Honda, I. et al. (2018) Neurosci. Lett. 665, 140. - Western blot analysis of mouse primary hippocampal culture lysate. Immunocytochemical staining of mouse cultured hippocampal neurons. Tested in Girk2-/- mice.
Marron Fernandez de Velasco, E. et al. (2017) Sci. Rep. 7, 1639. - Western blot analysis of mouse brain lysate. Tested in GIRK2-/- mice.
Koyrakh, L. et al. (2005) J. Neurosci. 25, 11468.
- Mouse brain lysate.
Tang, X. et al. (2018) Neuroscience 373, 52. - Mouse primary hippocampal culture lysate. Also tested in Girk2-/- mice.
Marron Fernandez de Velasco, E. et al. (2017) Sci. Rep. 7, 1639. - Mouse hippocampus lysate.
May, L.M. et al. (2017) Front. Neurosci. 11, 456. - Mouse perirhinal cortex lysate.
Roncace, V. et al. (2017) Neurobiol. Dis. 106, 89. - Mouse brain lysate.
Joshi, K. et al. (2016) Ann. Neurol. 80, 511. - Mouse brain lysate (1:200).
Kotecki, L. et al. (2015) J. Neurosci. 35, 7131. - Mouse brain lysate (1:1000).
Goodfellow, N.M. et al. (2014) J. Neurosci. 34, 6107. - Mouse brain lysate (1:200).
Hearing, M. et al. (2013) Neuron 80, 159. - Mouse brain lysate. Also tested in GIRK2-/- mice.
Koyrakh, L. et al. (2005) J. Neurosci. 25, 11468.
- Mouse brain membrane proteins (1:174).
Jelacic, T.M. et al. (2000) J. Biol. Chem. 275, 36211.
- Rat brain sections.
Arnold, E.C. et al. (2019) eNeuro 6, 0036-19. - Mouse brain sections (1:400). Also tested in GIRK2 conditional knockout mice.
Honda, I. et al. (2018) Neurosci. Lett. 665, 140. - Monkey brain sections.
Kikuchi, T. et al. (2017) Nature 548, 592. - Rat brain sections.
Niclis, J.C. et al. (2017) Stem Cell Reports 9, 868. - Mouse brain sections (1:100).
Stott, S.R.W. et al. (2017) PLoS ONE 12, e0171748. - Mouse brain sections (1:600).
Brandalise, F. et al. (2016) Eur. J. Neurosci. 43, 1460. - Rat brain sections (1:500).
Montandon, G. et al. (2016) Sci. Rep. 6, 32707. - Rat brain sections.
Seiler, S. et al. (2016) Front. Cell. Neurosci. 10, 87. - Mouse embryo sections.
Bye, C.R. et al. (2015) Proc. Natl. Acad. Sci. U.S.A. 112, E1946. - Mouse brain sections (1:1000).
Kotecki, L. et al. (2015) J. Neurosci. 35, 7131. - Mouse brain sections (1:600).
Bodea, G.O. et al. (2014) Development 141, 661. - Mouse and Rat DRG, sciatic nerve and skin sections (1:200).
Nockemann, D. et al. (2013) EMBO Mol. Med. 5, 1263. - Human skin sections (1:200).
Nockemann, D. et al. (2013) EMBO Mol. Med. 5, 1263. - Mouse spinal cords.
Marker, C.L. et al. (2004) J. Neurosci. 24, 2806.
- Mouse cultured hippocampal neurons. Also tested in Girk2-/- mice.
Marron Fernandez de Velasco, E. et al. (2017) Sci. Rep. 7, 1639. - hVM1 (human ventral mesencephalic neural stem cell line 1 (1:400).
Courtois, E.T. et al. (2010) J. Biol. Chem. 285, 9881.
- Paik, E.J. et al. (2018) Sci. Rep. 8, 804.
- Rivetti di val Cervo, P. et al. (2017) Nat. Biotechnol. 35, 444.
- Booker, S.A. et al. (2016) Cereb. Cortex 27, 2318.
- Robertson, D.N. et al. (2016) Methods 92, 19.
- Booker, S.A. et al. (2013) J. Neurosci. 33, 7961.
- Stott, S.R.W. et al. (2013) J. Neurosci. 33, 8022.
- Cooper, A. et al. (2012) Proc. Natl. Acad. Sci. U.S.A. 109, 2642.
- Ellisor, D. et al. (2012) Dev. Biol. 372, 249.
- Maity, B. et al. (2012) J. Biol. Chem. 287, 4972.
- Reyes, S. et al. (2012) J. Comp. Neurol. 520, 2591.
- Wydeven, N. et al. (2012) Proc. Natl. Acad. Sci. U.S.A. 109, 21492.
- Arora, D. et al. (2011) J. Neurosci. 31, 12251.
- Deleidi, M. et al. (2011) Stem Cells. 29, 1052.
- Lennington, J.B. et al. (2011) J. Neurosci. 31, 13078.
- Di Salvio, M. et al. (2010) Nat. Neurosci. 13, 1481.
- Grealish, S. et al. (2010) Brain 133, 482.
- Nassirpour, R. et al. (2010) J. Neurosci. 30, 13419.
- Wakita, S. et al. (2010) J. Neurosci. Methods 192, 83.
- Chung, H.J. et al. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 629.
- Friling, S. et al. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7613.
- Kolk, S.M. et al. (2009) J. Neurosci. 29, 12542.
- Iwanir, S. and Reuveny, E. (2008) Pflugers Arch. 456, 1097.
- Lignon, J.M. et al. (2008) Physiol. Genomics 33, 230.
- Labouèbe, G. et al. (2007) Nat. Neurosci. 10, 1559.
- Kulik, A. et al. (2006) J. Neurosci. 26, 4289.
- Marker, C.L. et al. (2006) J. Neurosci. 26, 12251.
- Huang, C.S. et al. (2005) Cell 123, 105.
- Marker, C.L. et al. (2005) J. Neurosci. 25, 3551.
- Thompson, L. et al. (2005) J. Neurosci. 25, 6467.
- Ehrlich, J.R. et al (2004) J. Physiol. 557, 583.
- Shankar, H. et al. (2004) Blood 104, 1335.
- Davila, V. et al. (2003) J. Neurosci. 23, 5693.
- Peleg, S. et al. (2002) Neuron 33, 87.
- Torrecilla, M. et al. (2002) J. Neurosci. 22, 4328.
- Inanobe, A. et al. (1999) J. Neurosci. 19, 1006.
- Pei, Q. et al. (1999) Neuroscience 90, 621.
- Kuzhikandathil, E.V. et al., (1998) Mol. Cell. Neurosci. 12, 390.
- Schein, J.C. et al. (1998) Dev. Biol. 204, 432.

