Overview
|
| Bioz Stars Product Rating | |
| The world's only objective ratings for scientific research products | |
| Mentions | |
| Recency | |
| View product page > | |
ω-Conotoxin GVIA (#C-300) is a highly pure, synthetic, and biologically active peptide toxin.
Applications
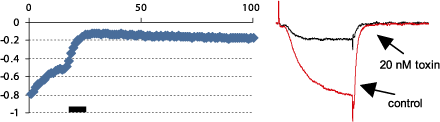 Alomone Labs ω-Conotoxin GVIA inhibits CaV2.2 heterologously expressed in Xenopus oocytes.Left: Time course of ω-Conotoxin GVIA (#C-300) action. Current amplitudes were plotted as a function of the stimulus number. Oocyte membrane potential was held at –100 mV and 100 ms stimulating pulse to 0 mV was delivered every 10 seconds. 20 nM ω-Conotoxin GVIA was perfused in the period marked by the bar. Right: Example traces of N-type currents before and during ω-Conotoxin GVIA application.
Alomone Labs ω-Conotoxin GVIA inhibits CaV2.2 heterologously expressed in Xenopus oocytes.Left: Time course of ω-Conotoxin GVIA (#C-300) action. Current amplitudes were plotted as a function of the stimulus number. Oocyte membrane potential was held at –100 mV and 100 ms stimulating pulse to 0 mV was delivered every 10 seconds. 20 nM ω-Conotoxin GVIA was perfused in the period marked by the bar. Right: Example traces of N-type currents before and during ω-Conotoxin GVIA application. Alomone Labs ω-Conotoxin GVIA blocks CaV2.2 channel in mouse Calyx of Held.Calyxes were whole-cell voltage-clamped at -80 mV. Traces were recorded in the presence of 1 mM CaCl2, pharmacological isolation of VGCC subtypes was performed in 2 mM CaCl2. 2 µM ω-Conotoxin GVIA (#C-300) was applied to selectively block CaV2.2.
Alomone Labs ω-Conotoxin GVIA blocks CaV2.2 channel in mouse Calyx of Held.Calyxes were whole-cell voltage-clamped at -80 mV. Traces were recorded in the presence of 1 mM CaCl2, pharmacological isolation of VGCC subtypes was performed in 2 mM CaCl2. 2 µM ω-Conotoxin GVIA (#C-300) was applied to selectively block CaV2.2.
Adapted from Lubbert, M. et al. (2017) eLife 6, e28412. with permission of eLife Sciences.
Citations (59)
 Knockdown of FMRP enhances synaptic vesicle exocytosis in presynaptic terminals of DRG neurons via CaV2.2 channels.A and B. vGpH response to 40 AP at 10 Hz from presynaptic terminals of DRG neurons transfected with Ctrl shRNA (A) or FMRP shRNA (B) before and after treatment with ω-Conotoxin GVIA (#C-300) and ω-Agatoxin IVA (#STA-500). Fluorescence intensities were normalized to the peak of a brief application of NH4Cl. C. Normalized vGpH responses to 40 AP at 10 Hz from presynaptic terminals of DRG neurons transfected with Ctrl shRNA (black-filled bar, 100±10.6%, n = 38) or FMRP shRNA (red open bar, 137.0±12.6%, n = 25, P = 0.027). ω-Conotoxin GVIA (ConoTx) reduces Ctrl shRNA and FMRP shRNA responses to a similar level (44.7±4.9%, n = 15 and 41.6±3.3%, n = 24, respectively). ω-Conotoxin GVIA and ω-Agatoxin IVA (AgaTx) application further reduces the responses: Ctrl shRNA = 17.3±3.2%, n = 38, and FMRP shRNA = 18.1±5.0%, n = 27. D. Average vGpH response to a 40 Hz stimulation for 30 s from presynaptic terminals of DRG neurons transfected with Ctrl shRNA (blackfilled squares) or FMRP shRNA (open red squares).
Knockdown of FMRP enhances synaptic vesicle exocytosis in presynaptic terminals of DRG neurons via CaV2.2 channels.A and B. vGpH response to 40 AP at 10 Hz from presynaptic terminals of DRG neurons transfected with Ctrl shRNA (A) or FMRP shRNA (B) before and after treatment with ω-Conotoxin GVIA (#C-300) and ω-Agatoxin IVA (#STA-500). Fluorescence intensities were normalized to the peak of a brief application of NH4Cl. C. Normalized vGpH responses to 40 AP at 10 Hz from presynaptic terminals of DRG neurons transfected with Ctrl shRNA (black-filled bar, 100±10.6%, n = 38) or FMRP shRNA (red open bar, 137.0±12.6%, n = 25, P = 0.027). ω-Conotoxin GVIA (ConoTx) reduces Ctrl shRNA and FMRP shRNA responses to a similar level (44.7±4.9%, n = 15 and 41.6±3.3%, n = 24, respectively). ω-Conotoxin GVIA and ω-Agatoxin IVA (AgaTx) application further reduces the responses: Ctrl shRNA = 17.3±3.2%, n = 38, and FMRP shRNA = 18.1±5.0%, n = 27. D. Average vGpH response to a 40 Hz stimulation for 30 s from presynaptic terminals of DRG neurons transfected with Ctrl shRNA (blackfilled squares) or FMRP shRNA (open red squares).
Adapted from Ferron, L. et al. (2014) with permission of Springer Nature.
- Rat INS-1 832/12 cells (whole-cell calcium currents).
Luan, C. et al. (2019) Commun. Biol. 2, 106. - Rat DRGs (patch clamp).
Moutal, A. et al. (2018) Neuroscience 381, 79.
- Torturo, C.L. et al. (2019) eNeuro 6, e0278.
- Lubbert, M. et al. (2017) eLife 6, e28412.
- Forostyak, O. et al. (2016) Stem Cell Res. 16, 622.
- Grimsley, C.A. et al. (2016) J. Neurophysiol. 116, 2550.
- Luster, B.R. et al. (2016) Physiol. Rep. 4, e12787.
- D’Onofrio, S. et al. (2016) Physiol. Rep. 4, e12740.
- Pennock, R.L. and Hentges, S.T. (2016) J. Neurophysiol. 115, 2376.
- Squecco, R. et al. (2016) Mol. Cell. Neurosci. 75, 50.
- D’Arco, M. et al. (2015) J. Neurosci. 35, 5891.
- Gan, K.J. et al. (2015) Mol. Biol. Cell 26, 1058.
- Lopez Soto, E.J. et al. (2015) J. Gen. Physiol. 146, 205.
- Cassidy, J.S. et al. (2014) Proc. Natl. Acad. Sci. U.S.A. 111, 8979.
- Ferron, L. et al. (2014) Nat. Commun. 5, 3628.
- Gomez-Sanchez, R. et al. (2014) Neurobiol. Dis. 62, 426.
- Hernandez-Gonzalez, O. et al. (2014) Purinergic Signal. 10, 269.
- Perez-Burgos, A. et al. (2014) FASEB J. 28, 3064.
- Tu, H. et al. (2014) Am. J. Physiol. 306, C132.
- Alamilla, J. and Gillespie, D.C. (2013) PLoS ONE 8, e75688.
- Amato, A. et al. (2013) Eur. J. Pharmacol. 718, 131.
- Eggers, E.D. et al. (2013) J. Neurophysiol. 110, 709.
- Izquierdo Serra, M. et al. (2013) Biochim. Biophys. Acta. 1830, 2853.
- Jones, S.L. and Stuart, G.J. (2013) J. Neurosci. 33, 19396.
- Kim, S.H. and Ryan, T.A. et al. (2013) J. Neurosci. 33, 8937.
- Lowe, M. et al. (2013) Exp. Physiol. 98, 1199.
- Radzicki, D. et al. (2013) J. Neurosci. 33, 9920.
- Shutov, L.P. et al. (2013) J. Physiol. 591, 2443.
- Sia, T.C. et al. (2013) Am. J. Physiol. 305, G933.
- Tzour, A. et al. (2013) J. Neuroendocrinol. 25, 76.
- Wedemeyer, C. et al. (2013) J. Neurosci. 33, 15477.
- Wen, H. et al. (2013) eLife 2, e01206.
- Wen, H. et al. (2013) J. Neurosci. 33, 7384.
- Abitbol, K. et al. (2012) J. Physiol. 590.13, 2977.
- Baillie, L.D. et al. (2012) Neuropharmacology 63, 362.
- Wang, S. et al. (2012) Cereb. Cortex 2, 584.
- Xiong, Q.J. et al. (2012) Am. J. Physiol. 303, C376.
- Alle, H. et al. (2011) J. Neurosci. 31, 8001.
- Martel, P. et al. (2011) PLoS ONE 6, e20402.
- Craviso, G.L. et al. (2010) Cell. Mol. Neurobiol. 30, 1259.
- Meunier, F.A. et al. (2010) J. Cell Sci. 123, 1131.
- Braun, M. et al. (2008) Diabetes 57, 1618.
Specifications
Technical Specifications
Biological Activity
- McCleskey, E.W. et al. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 4327.
- Lalo, U.V. et al. (2001) Brain Res. Bull. 54, 507.
Solubility and Storage
Scientific Background
ω-Conotoxin GVIA is a synthetic toxin originally isolated from the Conus geographus. ω-Conotoxin GVIA is a specific blocker of CaV2.2 Ca2+ channels. It specifically blocks N-type CaV channels by binding to the CaV2.2 α1 subunit (α1B) and its action is only partially reversible.2,3 In accordance, it inhibits synaptic transmission in many systems.4 It is also reported to antagonize P2X receptors.5 The toxin is used to specifically investigate CaV2.2 channel's contributions (by subtraction of the activity before and during perfusion of the toxin). Alternatively, it is used to eliminate the N-type channel contribution to highlight some other channel or enzyme activity. ω-Conotoxin GVIA was also used to purify the channel protein using immunoprecipitation techniques and to label and localize channels and synapses.6
- Olivera, B.M. et al. (1984) Biochemistry 23, 5087.
- McCleskey, E.W. et al. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 4327.
- Feng, Z.P. et al. (2001) J. Biol. Chem. 276, 15728.
- Kerr, L.M. and Yoshikami, D. (1984) Nature 308, 282.
- Lalo, U.V. et al. (2001) Brain Res. Bull. 54, 507.
- Pravettoni, E. et al. (2000) Dev. Biol. 227, 581.

